The Jupyter tool is a useful place to develop Python, R, or Octave code and analyses in a notebook style. Hub users can easily share their notebooks with other users by publishing notebooks as tools. A published Jupyter notebook enables other users to interact with the notebook, stepping through its cells and even changing them. When users run your published notebook, any changes they make to it will not persist.
Here we assume you are running: anaconda-6; debian10 container.
Python packages
Python has been extended to work with hundreds of specialized packages. For example, see the Anaconda package repo. A number of scientific Python packages are installed and accessible on Ghub.
Ghub uses Jupyter kernels to safely load needed Python packages. You can select a Jupyter kernel to set paths to a self-contained installation of specified packages, making them available in your notebook. This page will show you how to set access to Python packages from Jupyter Notebooks.
Note that we must install packages on Ghub to make them available as a kernel. Submit a ticket to request new packages or a new kernel. To find out what's available, refer to Kernel availability.
Selecting a kernel
New notebook
To select the kernel for a new notebook, start the Jupyter Deb 10 tool. In the upper right, select 'New', then the kernel you want from the kernel menu. You can now import and use the kernel's packages in your notebook.
Be sure to save the notebook after changing the kernel.
Existing notebook
If you need to change the kernel for an existing notebook, first open the notebook in the Jupyter Deb 10 tool.
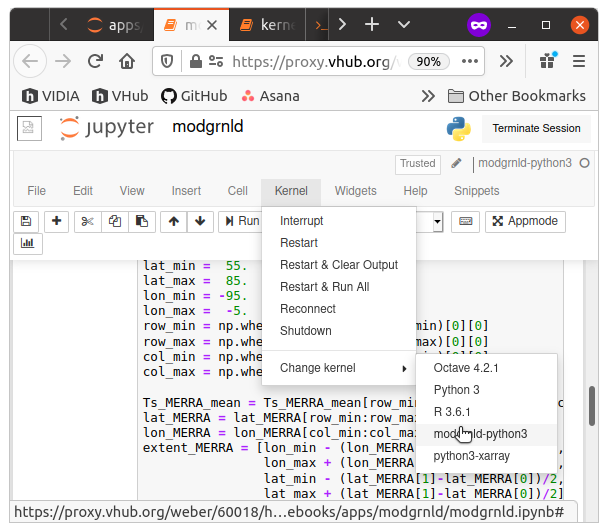
- If the notebook is already running, you must first shut it down by selecting Kernel: Shutdown from the menus.
- Then, you can select the kernel of your choice using the menu Kernel: Change Kernel: somekernel. After you have made the selection, check the displayed kernel name on the upper right of the notebook. It should match what you just selected.
- Finally, save the notebook, and your kernel choice will save along with it.
Kernel availability
How do we know what packages are available in what kernels?
- Check the conda env specification file associated with the kernel. Our conda env exports are kept up to date in a Git repo: https://github.com/ghub-gateway/ghub-envs
- Run conda commands to interrogate the packages. Read the next section for further information.
Using conda to list installed packages
The kernels we have created to support different sets of Python packages are based on conda environments ("envs"). You can interrogate these conda envs to list the packages a given kernel supports. This is general to Anaconda package manager (more is available here). Below are a few tips.
Note that creating a conda env is an administrator action. If you need a new env or additional packages, enter a ticket to request them.
Example
The kernel named modgrnld-python3 contains the following packages and their dependencies:
- matplotlib
- rasterio
- georaster
- hublib
- python 3.7
- netCDF4
- numpy
- pyproj
- scipy
What envs are available?
To access a conda env, first start a Workspace10 tool. On the command line, type the following commands to set the anaconda installation in your path:
source /etc/environ.sh use anaconda-6
Now, to show the names of available envs:
conda info --envs
Note that we may not have created kernels for all the available envs.
What packages are in this env?
If you have an env enabled currently, to list packages there, type:
conda list
Or for an arbitrary env, someenv:
conda list -n <someenv>
Export current conda env
To export a list of the packages and versions installed in the env to a text-based .yml file:
conda activate <envname> conda env export > <filename>.yml